Metabolic Engineering
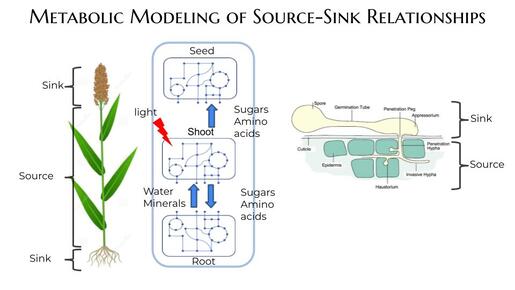
We combine computational modeling, genetics, genomics, biochemistry, synthetic biology, and bioinformatics to model genome-scale metabolic networks of cells, organ systems, and organisms to understand, predict, and engineer metabolic traits. Examples of systems we study include source-sink relationships within a plant to maximize biomass production under suboptimal conditions, plant-pathogen relationships to identify the sources of metabolic dependency, and microbe-microbe interactions within a microbial community in the fruit fly gut. We also use omics data to decipher the rules of transcriptional regulation of metabolism and have discovered novel epigenetic and transcription factor-mediated regulatory mechanisms. Finally, we combine computational and synthetic biology methods to discover new biosynthetic pathways that synthesize natural products in plants.
Learn more about some of our federally funded projects:
Learn more about some of our federally funded projects:
- Setaria for Biofuels: The objective of this project is to utilize genomic, computational, and engineering tools to begin the genetic dissection of drought and density response in S. viridis. Through the use of S. viridis, we hope to discover the molecular mechanisms that underlie this response and in doing so identify candidate genes and pathways for improving the closely related feedstock grasses.
- Plant Metabolic Network: This network currently houses one multi-species reference database called PlantCyc and 126 species/taxon-specific databases. PlantCyc provides access to manually curated or reviewed information about shared and unique metabolic pathways present in over 500 plant species.
- Sorghum Metabolic Atlas: This project aims to holistically decipher plant metabolic networks to tackle the impending challenges of changing climate, food security, and availability of sustainable energy sources. The research team will create a high-quality subcellular map of small molecule metabolism in Sorghum and Brachypodium.
- Pennycress Metabolic Modeling: Pennycress is under development as a winter annual oilseed bioenergy crop for the 80 million-acre U.S. Midwest Corn Belt and other temperate regions including the Pacific Northwest. This project will deliver speed-breeding methods to facilitate the introduction of superior allelic variants into a wide range of commercial pennycress varieties to precisely adapt them to the desired local environments.
- Sorghum for Biofuels: This project aims to develop novel technologies and methodologies to redesign the bioenergy feedstock Sorghum bicolor to enhance water use efficiency and photosynthetic efficiencies, using Setaria as a model. Improving these two traits will make sorghum and other panicoid grasses (e.g. Miscanthus, switchgrass) more productive on marginal soils.






