Data-Driven Discovery
Over 25-75% of protein-encoding genes in all sequenced genomes are not similar to anything with known molecular functions. We develop approaches to systematically characterize these ‘unknown’ genes.
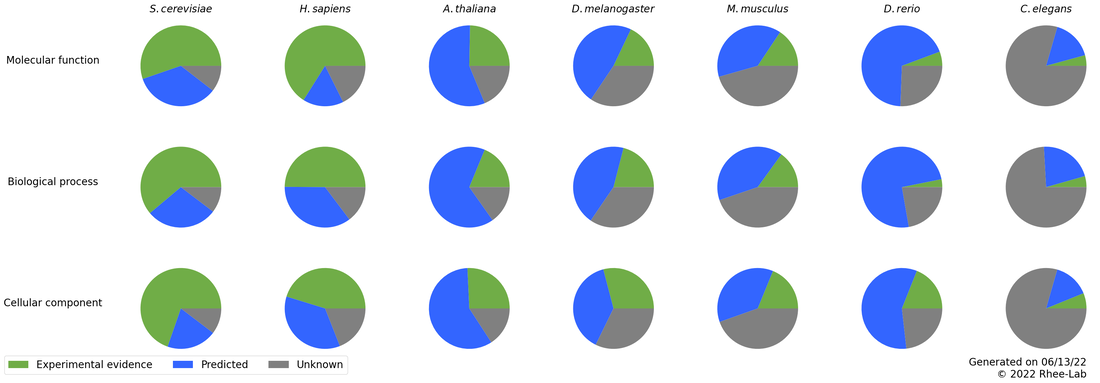
Graphical overview of the status of genome function annotations for model species and select bioenergy & food crops
This part of our research program is the riskiest, but we are learning the newest biology from this approach. One example is a novel gene that we call CHIQUITA1, which is a negative growth regulator that attenuates cell division rates early in development. In the absence of the protein, cell division is accelerated during early development; organs switch from proliferation to differentiation earlier than the wild type, consigning the mutant plants to become dwarf at maturity. This is a holy-grail trait --early vigor combined with late onset dwarfism-- that farmers have been looking for since the Green Revolution. Another example is a gene we named FLOE1 (after the second movement of Philip Glass’s Glassworks), which acts as a water sensor and regulates seed germination. Upon exposure to water, the protein changes its physical nature from a glassy state to liquid droplets, which we believe changes the viscosity of the cell cytoplasm, thereby allowing metabolic activities and the germination process to start. We are currently translating these discoveries into oil crops and sorghum. Considering that these are the first two genes of the ‘unknown’ genes we have been characterizing, I am confident that this dark matter of the genome in plants is a gold mine for the riskphilic, creative, and optimistic.